Download 140128 学位論文表紙
Transcript
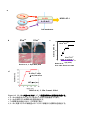
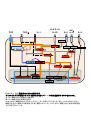
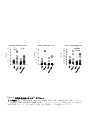
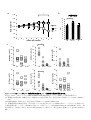
IL-1 Th17 (The mechanism of Th17 cell development induced by excess IL-1 signaling) 23 12 (Il1rn-/-) IL-1 IL-17 Th17 TGF-β IL-6 Il1rn-/- IL-6 Il1rn-/-Il6-/IL-6 naïve CD4+T Il6-/- naïve T Th17 TCR TGF-β IL-1 TGF-β+IL-1+IL-21 Th17 IL-21 TGF+IL-21 TGF-β IL-1 Th17 Th17 Th17 Th17 IL-1 IL-1 TGF-β TGF-β Foxp3 Th17 IL-6 IL-1 IL-1 Nfkbiz IL-6 Batf Th17 Th17 2 IL-21 Abstract IL-1 receptor antagonist-deficient (Il1rn-/-) mice spontaneously develop autoimmune arthritis and that IL-17 is crucial for the onset of the disease. Although many studies have shown that the Th17 cell differentiation is dependent on TGF-β and IL-6, we found that IL-6 deficiency does not affect the development of arthritis in Il1rn-/- mice at all. In vivo, Th17 cells developed normally in Il1rn-/-Il6-/- mice. To elucidate the mechanisms of IL-6 independent Th17 cell differentiation, we purified naïve CD4+ T cells from Il6-/- mice and cultured with TCR stimulation, TGF-β, IL-1 and/or IL-21. We found that naïve CD4+ cells efficiently differentiated into Th17 cells when cultured with TGF-β, IL-21 and IL-1 than the combination of TGF-β and IL-21, however, these cells did not differentiate with TGF-β and IL-1. In addition, IL-1 maintained Th17 lineage synergistically with TGF-β. Interestingly, we found that IL-1 inhibited TGF-β-induced Foxp3 expression to promote Th17 cell differentiation in a proliferation, IL-6 and IL-21 independent manner. Furthermore, IL-1 augments Th17 transcription factors expression such as Nfkbiz and Batf. These results indicate that IL-1 plays an important role in the Th17 cell differentiation in an IL-6 independent manner. 3 1-1 2-3 1-2 5-8 1-3 9-17 1-4 18-27 1-5 28-37 1-6 38 39-41 42-51 1-7 52-76 4 (McInnes and Schett, 2007) (McInnes and Schett, 2007) IL-6 IL-6 CIA HTLV-1 transgenic (Tg) SKG (Figure 0-1a)(Ishihara et al., 2004; Iwakura et al., 2008) IL-6R (Figure 0-1b) (Jones et al., 2010; Nishimoto et al., 2009) IL-1 (Il1rn-/- mouse) IL-1 IL-1Ra (Horai et al., 2000)( Figure 0-2a b) IL-1 CIA HTLV-I Tg Figure 0-1a SKG K/BxN (Iwakura et al., 2008) IL-1 IL-1Ra 2002; Furst, 2004) Th17 IL-1 (Cohen et al., IL-1 IL-17 et al., 2009)(Figure 0-3) Th17 IL-6 CD4+ CIA 5 T (Korn EAE (experimental autoimmune encephalomyelitis) (Iwakura et al., 2011) IL-17 Th17 (Genovese et al., 2010; Hueber et al., 2010) naïve T TGF β IL-6 2007) TGF β Foxp3 IL-21 (Figure 0-3)(Zhou et al., Th17 retinoic acid receptor-related orphan receptor (ROR)-γt Zhou et al., 2008) IL-6 IL-21 IL-23 IL-17 STAT3 RORγt RORγt (Ichiyama et al., 2008; (Figure 0-4) STAT3 IL-17 IRF-4 IL-17 (Hirahara et al., 2010) STAT3 Nfkbiz Hif1a Ahr Th17 (Figure 0-4) (Dang et al., 2011; Kimura et al., 2008; Okamoto et al., 2010) STAT3 CD4+T Th17 IL-21 STAT3 IL-23R Th17 (Durant et al., 2010; Zhou and Littman, 2009) IL-23 Th17 IL-17A IL-17F IL-22 IL-1β IL-17 IL-1 PKCθ T IL-17 al., 2009; Sutton et al., 2006; Yang et al., 2008a) (Chung et al., 2009) IL-1 (Kryczek et al., 2007) IL-6 IL-1R1 mTOR PI3K NF-κB IL-2 IL-6 IRF-4 Th17 SIGIRR Th17 6 (Korn et IL-1R (Gulen et al., 2010) Foxp3 IL-1 Th17 T(Treg) IRF-4 Blimp-1 IL-10 CTLA-4 ICOS GITR Treg (Cretney et al., 2011; de Lafaille and Lafaille, 2009) Foxp3+T Induced Foxp3+ Treg:iTreg (de Lafaille and Lafaille, 2009) TCR IL-2 TGF-β Th17 Foxp3 TGF β TGF-β Foxp3 IL-6 Foxp3 RORγt IL-21 Treg RORγt RORγt IL-23 Th17 Foxp3 Th17 RORγt (Ahern et al., 2010; Ichiyama et al., 2008; Zhou et al., 2008)(Figure 0-5) Th17 Treg (Bettelli et al., 2006) (Figure 0-2 and Figure 0-3) Foxp3 RORγt IL-17 Il1rn-/- IL-6 Il1rn-/-Il6-/- IL-6 / Il1rn-/-Il6-/- Il1rn-/- IL-1 IL-6 IL-1 IL-1 IL-6 IL-6 Th17 Th17 Th17 Th17 IL-1 TGF-β 7 Foxp3 Th17 Th17 IL-6 IL-1 8 Il1rn-/- (Il1rntm1Yiw) Il1ab-/- (Il1atm1Yiw/Il1btm1Yiw) (Horai et al., 1998; Horai et al., 2000) Il1rn-/-Il6-/(Il6tm1Kopf M. Kopf Il1rn-/- Il6-/- )(Kopf et al., 1994) Il1r1-/- Immunex Corporation Rag2-/- (Glaccum et al., 1997) (Shinkai et al., 1992) 8 BALB/cA ( Il1rn-/- ) Il1rn-/-Il6-/MyD88-/- (MyD88tm1Aki) C57BL/6J (Adachi et al., 1998) C57BL/6J (Nihon SLC; Shizuoka, Japan) BALB/cA (CLEA Japan, Inc.) SPF (specific pathogen-free conditions) 10% FBS RPMI1640: 50 U/ml penicillin invitrogen ) 50 mM 2-mercaptoethanol (GIBCO; 50 µg/ml streptomycin 10% FBS (w/v) RPMI1640 (nakarai tesque; Kyoto, Japan) 9 10% FBS RPMI1640 FBS Hemolysis Buffer: 1.4 M NH4Cl T Th17 0.45 mm Tris-HCl (pH7.2) 170 mM 17mM Tris-HCl (pH7.2) mM NH4Cl 140 Hemolysis Buffer MACS Buffer: ( ) g 1L 5 0.45 µm MACS Buffer FACS Buffer: HBSS (Hanks' Balanced Salt Solutions) (Nissui, Tokyo) FBS 0.1% NaN3(Nakarai tesque, Tokyo) 2% FACS Buffer anti-CD3 anti-CD3(clone 145-2C11; ) 48 well/ Flat Bottom (Asahi glass) PBS anti-IL-4 37 PBS 4 2 anti-IFN-γ anti-IL-4 (clone 11B11) anti-IFN-γ 11B11 (clone R4-6A2) CeLLine BD Biosciences Freund’s Incomplete Adjuvant 0.5 ml BALB/cA-nu/nu 10 PIERCE Biotechnology R4-6A2 1 106 cells/ mouse 1~2 1 37 2000rpm Cleanascite BIOTECH SUPPORT GROUP 0.45 µm 11B11 R4-6A2 HiTrap Protein G GE Healthcare Slide-A-Lyzer 10,000 MWCO Dialysis Cassette PIERCE; Thermo Fisher Scientific PBS BCA Protein Assay Reagent PIERCE; Thermo Fisher Scientific CD4+ Il6-/- 10% FBS RPMI1640 cm 3.5 (FALCON; BECTON DICKINSON) (on ice) 108 µm 1300 rpm MACS Buffer MACS Buffer Micro beads CD4 (Miltenyi Biotec GmbH; Bergisch, German) 15 Auto MACS possel program (Miltenyi Biotec GmbH; Bergisch, German) CD4+ anti-CD3 5 105 cells (clone 145-2C11) GLASS; Tokyo, Japan) San Jose, CA, USA) 500 µl/48well 4 µg/ml 48well/ Flat Bottom (ASAHI anti-CD28 (1 µg/ml) (clone 37.51; Biolegend; 10 µg/ml anti-IFN-γ (clone R4-6A2) 10 µg/ml anti-IL-4 (clone X-VIVOTM 20 (Lonza; Basel, Switzerland) 11B11) CD4+ 5 37 /5% CO2 10 ng/ml rmIL-1α (PeproTech; Rocky Hill, NJ, USA) 10 ng/ml rmIL-1β (PeproTech) 11 naïve T T Naive CD4+ T Il1rn-/- BALB/cA BALB/cA Il6-/- Il1rn-/-Il6-/- Il1ab-/- C57BL/6J / MyD88+/- MyD88-/- 1300 rpm Hemolysis Buffer (1~2 ml) 5 10% FBS RPMI1640 MACS Buffer anti-mouse B220 anti-mouse CD8α anti-mouse CD11b Biosciences; San Jose, CA, USA) BlueTM- anti-mouse DX5 anti-mouse Ter119 PE-Cy7 anti-mouse CD4 (Biolegend) anti-mouse CD62L (Biolegend) PE San Diego, CA, USA) 4 Micro beads (BD anti-mouse CD25 20 Pacific (eBioscience, MACS Buffer anti-Biotin 4 15 deplete program AutoMACS (Miltenyi Biotec GmbH) selection negative FACS Aria(BD bioscience) CD4+CD62L+CD25- naïve T FACS Aria FACS naïve T X-VIVOTM 20 (Lonza) naïve CD4+ T 4 µg/ml anti-CD3 (clone 145-2C11) 1 µg/ml anti-CD28 (clone 37.51; Biolegend) 96 well Flat Bottom(FALCON; BECTON DICKINSON well 11B11) (PeproTech) ) 10 µg/ml anti-IFN-γ (clone R4-6A2) 2x105 cells 250 µl/ 10 µg/ml anti-IL-4 (clone 3 ng/ml CHO cell derived rhTGF-β1 (PeproTech), 40 ng/ml rmIL-6 10 ng/ml rmIL-1α (PeproTech) 12 10 ng/ml rmIL-1β (PeproTech) 100 ng/ml rmIL-21 (PeproTech) Laboratories 10 ng/ml rmIL-23(R&D systems) ) 50 ng/ml Rapamycin (LC IL-21R Subunit/Fc Chimera R&D systems (Minneapolis, MN, USA) CFSE T (1x106 to 1x107 cells/ml) 5 µM CFSE (5- and 6-carboxyfluorescein diacetate succinimidyl ester; invitrogen; Carlsbad , CA, USA) / PBS 6 37 8x105 /ml FACS CantoIITM (BD Biosciences) FlowJo software (TreeStar; Ashland, OR, USA) 50 ng/ml PMA (Sigma) 500 ng/ml ionomycin (Sigma) 2 µM monensin (Sigma) 37 5% CO2 anti-FcγRII/III receptor mAb (2.4G2) 5 4 10 2 µg/ml 7AAD (Sigma) FACS Buffer 4% paraformaldehyde 200 µl 20 permeabilization buffer (0.1% saponin containing FACS solution) 2 2 FACS Buffer 40 70 µm 13 /4 FACSCalibur™ FACSCantII APC- anti-CD4 (BD Bioscience) PE-Cy7- (eBiosciece) APC-Cy7PE- anti-CD4 (Biolegend) anti-B220 (Biolegend) FITC- FITC- anti-TCRβ (Biolegend) anti-IL-1R1 (Biolegend) Pacific Blue- : FITC or PE or anti-IFN-γ (Biolegend), FITC or APC or Pacific Blue- (Biolegend), APC or PE(eBioscience) anti-CD8a anti-Foxp3 (BD Biosciences), APC- Foxp3 anti-IL-17 RORγ(t) Foxp3 staining Kit (eBioscience) STAT3 Pacific Blue- BD phosflow Technology STAT3 (pY705) (BD bioscience ) PBS 1.5 cm PBS EDTA/10% FCS RPMI 20 ml 20 37 5 mM 50 ml ( ) 1~2 cm 5 mM EDTA/10% FCS RPMI 20 ml EDTA PBS 50 ml 20 10% FCS/2 mg/ml Collagenase/ 5 ml RPMI 37 Collagenase Collagenase 15 ml 200 µm 1300 rpm 5 14 45% GE healthcare 1:9 10% FBS RPMI1640 20 r.t 10xPBS 2200 rpm 10% FBS RPMI1640 2 lamina proprial lymphocytes; LPL PMA/ionomycin CD4+CD45RBhiCD25BALB/c Il1r1-/- BALB/c Il17a-/CD4+CD45RBhiCD25- naïve T FACS Aria 2 2x106 cells/ml PBS (BALB/c 200 µl PBS (4x105 cells/mouse) Rag2-/- ) MCM 4% (w/v) (Nissui; Tokyo, Japan) 2 ml 0.5 % EDTA PBS 10 % FBS RPMI1640 Japana 4% none treated 10 5x106 cells/ml 37 3 PBS glass) 15 5% CO2 Asahi glass, Tokyo, 4 (Asahi 5x105 cells/ml 24 well-plate (Asahi glass) 10% FBS RPMI1640 1 ml 2 LPS 5 µg/ml Zymosan 50 µg/ml 2 MCM LPS (Escherichia coli O55:B5) cerevisiae ) T Zymosan A (Saccharomyces SIGMA (St Louis, MO, USA) Real-Time RT-PCR RNA GenEluteTM Mammalian Total RNA Miniprep Kit (Sigma) RNA the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Inc.; CA, USA) real-time RT PCRs SYBER® Premix Ex TaqTM (TaKaRa; Shiga, Japan) iCyclerTM System (Bio-Rad; Hercules, CA, USA) 50 3:00 95 2:00 (95 0:15 60 1:00)x44 70 89.2 0.5 : Gapdh 5’-TTCACCACCATGGAGAAGGC-3’ 5’-GGCATGGACTGTGGTCATGA-3’ Foxp3 5’-AGAAGCTGGGAGCTATGCAG-3’ 5’-TACTGGTGGCTACGATGCAG-3’; Il17a 5’-CTCCAGAAGGCCCTCAGACTAC-3’ 5’-GGGTCTTCATTGCGGTGG-3’ Rorc 5’-AGCAGTGTAATGTGGCCTAC-3’ 5’-GCACTTCTGCATGTAGACTG-3’ Il1r1 5’-ACCTTCCCACAGCGGCTCCACATT-3’ 5’-TTGTCAAGAAGCAGAGGTTTACAG-3’ Gata3 5’-CTTATCAAGCCCAAGCGAAG-3’ 5’-CATTAGCGTTCCTCCTCCAG-3’ Tbx21 5’-GGTGTCTGGGAAGCTGAGAG-3’ 5’-CCACATCCACAAACATCCTG-3’ 16 Il21 5’-GCCAGATCGCCTCCTGATTA-3’ 5’-CATGCTCACAGTGCCCCTTT-3’ Il22 5’-TGACGACCAGAACATCCAGA-3’ 5’-AGCTTCTTCTCGCTCAGACG-3’ Batf 5’-CCAGAAGAGCCGACAGAGAC-3’ 5’-GAGCTGCGTTCTGTTTCTCC-3’ Nfkbiz 5’-CCTCCGATTTCTCCTCCACT-3’ 5’-GTTCTTCACGCGAACACCTT-3’ 1 4 0; 1; 2; t- 3; P<0.05 17 Il1rn-/- IL-6 Th17 Il1rn-/IL-1β IL-6 IL-17 TNF-α Il1rn-/- (Horai et al., 2004; Nakae et al., 2003) Il1rn-/-Il6-/- IL-6 Il1rn-/-Il6-/- Il1rn-/- (Figure 1a) Il1rn-/- IL-6 B220+ CD4 Il1rn-/-Il6-/- CD8+ Il1rn-/- (Figure 2) Il1rn-/-Il6-/CD4+ Th17 Th17 Il1rn-/- Il1rn-/- (Figure 1b 3) Th17 IL-6 IL-1 IL-6 Il1rn-/-Il6-/IL-6 Th17 Th17 IL-1 Th17 Il6-/- Th17 Il1rn-/-Il6-/- CD4+ Il6-/- IL-1 IL-1 IL-6 (Figure 4) (Kryczek et al., 2007) 18 CD4+IL-17+ in vitro CD4+ IL-6 IL-21 TGF-β Il1rn-/-Il6-/- Th17 IL-6 Th17 IL-21 Il1rn-/- Il6-/- Il1rn-/-Il6-/IL-21 mRNA Il1rn-/- PCR Il1rn-/-Il6-/- Il6-/- Il21 (Figure 5) IL-1 IL-21 IL-1 IL-21 Th17 naïve CD4+ T T in vitro naïve T (CD4+CD62LhiCD25- Th17 IL-1 CD4 /TGF-β TGF-β+IL-6 /IL-1+TGF-β TGF-β+IL-21 Th17 Th17 Th17 (Figure 6a c Figure 7) Th17 IL-1 IL-17 Figure 6c naïve CD4+ T naïve CD4+ Th17 T IL-1R1 FACS IL-6 IL-1 IL-22 IL-21 IL-1 ) naïve T PCR IL-1R1 IL-21 IL-1R1 (Figure 6b c Figure 8) Th17 IL-1 IL-6 IL-21 IL-1R1 naïve T Th17 Th17 19 IL-1R IL-1 Th17 in vitro Th17 Th1 TGF-β (Lee et al., 2009) IL-1 IL-17 IFN-γ IL-17A IL-17F Th17 Th17 (Chung et al., 2009) Th17 IL-1 TGF-β+IL-21 round naïve T 2nd 5 10 3rd round (Lee et al., 2009) TGF-β TGF-β IL-1 (Figure 9) IL-1 IL-1 TGF-β Th17 IL-17 Th17 IL-21 IL-1 TGF-β Th17 IL-17 IL-17 exogenous Th17 IL-17 Th17 Th17 IL-1 Th17 TGF-β+IL-21 naïve T Th17 IL-1 naïve T IL-1 IL-1 IL-6 IL-6 IL-1 IL-17A Th17 IL-6 IL-22 20 mRNA Th17 T (Figure 10a) IL-1 IL-21 IL-1 Th17 Rorc IL-1 IL-1 Foxp3 Nfkbiz Batf Foxp3 (Figure 10b) Foxp3 IL-1 Foxp3 Th17 Th IL-1 T Tbx21 Th1 Th2 Th17 Treg Figure 9 Gata3 Rorc Il17a TGF-β Gata3 Rorc Foxp3 mRNA 2nd round IL-1 Tbx21 IL-1 IL-1 Foxp3 (Figure 10c) IL-1 Foxp3 FACS Foxp3 (Figure 10d) Foxp3 IL-1 IL-1 MyD88 naïve T CD3 MyD88+/- FACS Th17 CD28 (TGF-β+IL-21) TGF-β IL-1 2 MyD88 (Figure 10e) IL-1 IL-1 MyD88 Th17 IL-17+ Foxp3 Foxp3 21 Foxp3 Foxp3 Foxp3 IL-1R1 (Figure 11) Th17 4 FACS T MyD88-/- IL-1 IL-1 T (Sims and Smith, 2010) IL-1 Foxp3- Foxp3+ Foxp3 naïve T Th17 IL-1 Foxp3 MFI PE- TGF-β/IL-1/IL-21 T TGF-β/IL-21 Foxp3+ (Figure 12a b) TGF-β iTreg 96 Foxp3 Foxp3 CFSE TGF-β+IL-21 TGF-β+IL-1+IL-21 TGF-β+IL-6 IL-1 Foxp3 IL-1 Foxp3 IL-1 Foxp3 IL-1 Foxp3 IL-6 (Figure 12c) IL-21 et al., 2007) IL-6 STAT3 IL-1 IL-21 Foxp3 Foxp3 (Huehn et al., 2009; Wei IL-6 IL-6 naïve T IL-1 (Figure 13a) TGF-β+IL-21 IL-21R-Fc Foxp3 IL-21 IL-6 Th17 Foxp3 IL-1 Foxp3- IL-1 T IL-21 22 IL-17 IL-1 Il6-/- naïve T IL-21 IL-6 (Figure 13b) IL-21 autocrine IL-21 IL-1 Foxp3 IL-1 IL-21R-Fc (20 µg/ml) (Figure 13c) Foxp3 IL-21R-Fc Foxp3 IL-21 STAT3(pY705) IL-1 STAT3 IL-21R-Fc STAT3 IL-21R-Fc Foxp3 IL-1 IL-21 IL-21 IL-1 STAT3 Foxp3 (Figure 13d) IL-1 Foxp3 IL-6 IL-21 IL-1 T IL-6 IL-6 Foxp3 IL-1Ra IL-1 IL-1 Foxp3 (Horai et al., 2004; Li and He, 2006; Il6-/- Van Kooten et al., 1991) CD4+T T Il1rn-/-Il6-/- Th17 (TGF-β+IL-21) anti-CD3/anti-CD28/IL-1 2 Foxp3 Foxp3 TGF-β Foxp3 Il6-/- TGF β naïve T Foxp3+ Th17 FACS 23 Il1ab-/IL-1 IL-6 IL-1 Il1ab-/- IL-17 Il1rn-/-Il6-/- Figure 13e Foxp3 naïve 4 IL-1 Foxp3 IL-21 Il1ab-/- Il1ab-/- naïve T T IL-6 Figure 14 IL-1Ra Th17 T IL-1 IL-1 IL-6 IL-21 Foxp3 IL-6 IL-1 Foxp3 Th17 IL-1 IL-21 IL-21 Foxp3 IL-1 IL-21 IL-1 IL-21 / IL-21 Il6-/-naïve T TCR IL-21R-Fc IL-1 TGF-β/IL-21 2nd round 2nd round 24 mRNA Real time PCR Rorc Il21 IL-1 IL-21 15 IL-1 Figure IL-21 Foxp3 Batf Il17 Il22 IL-21 IL-1 Foxp3 IL-1 IL-21 IL-21 Nfkbiz IL-1 Nfkbiz IL-1 IL-21 IL-1R1 IL-21 IL-1 IL-21 IL-1R1 Th17 IL-1R1 24 IL-1 IL-1Ra IL-21 Th17 Th17 Th17 IL-6 IL-6 Th17 (Ivanov et al., 2006) Il1rn-/- (Abdollahi-Roodsaz et al., 2008) Il1rn-/- Il1rn-/-Il6-/- Th17 Il1rn-/Th1 Il6-/- Il1rn-/-Il6-/- Treg Il6-/- Th17 Th17 (Figure 16) Il1rn-/- Th17 Th17 Treg IL-6 IL-1Ra in vivo In vivo Il1rn-/- Treg IL-1 IL-1 Th17 Th17 Il1r1-/- CD4+CD45RBhiCD25CD4+CD45RBhiCD25- Th17 iTreg iTreg Rag2-/Rag2-/- naïve T 25 Th17 Foxp3+ iTreg (Ahern et al., 2010) の Il17a-/- CD4+CD45RBhiCD25- Il1r1-/- CD4+CD45RBhiCD25- (Figure Il1r1-/- 17a) (Figure 17b) mesenteric lymph nodes Th1 Th1 Th17 Treg Treg Th17 Figure 17c Th17 T IL-17 IL-1 IL-1 IL-6 Th17 LPS zymosan Il6-/-MCM (MCM) LPS CD4+ Th17 Il1rn-/- TLR4 (Abdollahi-Roodsaz et al., 2008) zymosan Th17 (LeibundGut-Landmann et al., 2007; Veldhoen et al., 2006) CD3 CD28 IL-4 IFN-γ MCM Il6-/- FACS (Figure 18a) CD4+ Th17 MCM LPL zymosan Il1rn-/-Il6-/- Th17 MCM Il6-/- Il6-/- MCM Th17 IL-1 IL 6 naïve T 26 TGF β Th17 Il6-/- MCM naïve T Th17 Il6-/- (Figure 18b) Th17 MCM Th17 Foxp3 Th17 Th17 IL-6 27 HTLV-1 Tg SKG CIA IL-6 human TNF-α Tg (CAIA) T IL-6 (Iwakura et al., 2008) IL-6 IL-17 IL-6 in vivo in vitro Il1rn-/- Th17 IL-17 Il1rn-/-Il6-/- T Il1rn-/Il1rn-/-Il6-/- (Horai et al., 2004; Nakajima et al., 2010) Th17 Il1rn-/- IL-6 IL-1 Foxp3 TGF-β IL-1 Th17 Th17 Th17 Th17 Th17 IL-1 β TGF Th17 IL-6 1; IL-1R IL-21 IL-1R1 naïve T IL-1 Foxp3 naïve T TGF β+IL-21 naïve T Il1rn-/- naïve T IL-1R1 IL-21 Il1rn-/IL-1 IL-6 IL-21 IL-6 T STAT3 28 IL-1Ra IL-21 IL-1R1 IL-1R1 (Chung et al., 2009) IL-21 IL-21 IL-1R1 TGF β STAT3 IL-1R1 IL-21 mRNA CD4 FACS IL-21 (data not shown) CD4+ IL-21 CD4+ IL-21 NKT CD4+Th17 CD4 Tfh CD4+ (Monteleone et al., 2008) IL-21 IL-21 IL-6 IL-12 IL-21 (Nakayamada et al., 2011; Schmitt et al., 2009) IL-21 al., 2012) (Puga et IL-21 (Bubier et al., 2009; Spolski and Leonard, 2010) Th17 Th17 IL-1 IL-21 IL-1R1 IL-1 (Figure 6) IL-17 IL-1R1 STAT3 IL-1R1 IL-1R1 Th17 IL-1 Nfkbiz IL-1R1 29 Batf IL-1R1 2; Foxp3 Th17 IL-6 STAT3 STAT3 IL-17 TGF β Th17 RORγt Th17 IL-21 IL-6 Foxp3 TGF β et al., 2007) IL-1 IL-6 IL-6 (Yang et al., 2008b) (Korn Th17 IL-1 IL-1 IL-1 T RORα Th17 IL-21 Foxp3 Foxp3 Th17 STAT3 IL-21 IL-6 Foxp3 IL-6 IL-21R-Fc IL-21 IL-1 IL-21R-Fc IL-21 IL-21 IL-21 IL-1 IL-21R-Fc IL-21R-Fc Fc IL-6 IL-21 Foxp3 IL-1 Foxp3 IL-21 Foxp3 IL-1 Foxp3+ Foxp3 IL-1α/β IL-6 Foxp3- Foxp3 IL-1 Foxp3 Foxp3 EGFP IL-1 Foxp3 30 IL-2 Th17 IL-1 (Huehn et al., 2009) IL-2 Th17 STAT5 Foxp3 IL-1 (Huehn et al., 2009) IL-2 Foxp3 IL-1 Akt-mTOR mTOR (Delgoffe et al., 2011) Smad2 IL-1 Smad7 Foxp3 TGF βR2 TGF β (Bauge et al., 2007; Ishida et al., 2006) IL-1 Foxp3 Rapamycin mTOR Foxp3+ Figure 19 IL-1 Foxp3 mTOR MFI IL-1 IL-1 Foxp3 IL-1 Foxp3 Foxp3 (Huehn et al., 2009) IL-1 Nfkbiz IL-1 Foxp3 Batf Th17 Il17a Il22 IL-6 IL-1 IL-21 Th17 (Chung et al., 2009; Guo et al., 2009) Th17 IL-1 TGF-β Th17 (Lee et al., 2009) ILStriteski et al. 31 TGF-β Th17 IL-1 IL-23 Th17 (Stritesky et al., 2008) IL-1 Th17 Th17 IL-1R1 Rag2-/- naïve T Th17 in vivo Foxp3 (Figure 17) IL-1 Th17 Foxp3+ Foxp3+ iTreg Il1r1-/- Foxp3+ T IL-1 IL-6 IL-21 nTreg Foxp3 IL-17 IL-1 IL-17 2008b) Foxp3 (Chung et al., 2009; Yang et al., IL-1 Foxp3 Foxp3 IL-1 nTreg naïve T IL-1 IL-6 Foxp3+ iTreg Th17 Foxp3+ Foxp3 IL-1R1 Foxp3+ Th17 IL-21 Th17 IL-1R1 Foxp3+ nTreg Foxp3 (de Lafaille and Lafaille, 2009) Foxp3 IL-1 32 Foxp3 T (Zhou et al., 2009) 3; Il1rn-/- IL-6 TNF-α (Tak and Kalden, 2011) al., 2009) IL-1 IL-6R (Jones et al., 2010; Nishimoto et (Cohen et al., 2002; Furst, 2004) IL-17 (Genovese et al., 2010; Hueber et al., 2010) anti-IL-6R anti-IL-6R 30% Il1rn-/- (Jones et al., 2010) IL-6 Il1rn-/- IL-6 IL-17 (Nakajima et al., 2010) psoriatic arthritis (Ramonda et al., 2011) (Ramonda et al., 2011) Il1rn-/(Alexander et al., 2012) IL-6 IL-17 IL-1 33 Il1rn-/TNF-α 4; 4-1; Il1rn-/Il1rn-/- Il1rn-/Il1rn-/- B B B IL-21 anti-CD20 B B Il1rn-/- IL-21 Th17 IL-21 Il1rn-/- Il1rn-/- Autoimmune disease Auto inflammatory disease (Dinarello, 2009) Human IL-1Ra Auto inflammatory disease (Aksentijevich et al., 2009; Reddy et al., 2009) Auto inflammatory disease TCR Il1rn-/)/Rag2-/- TCR Tg Il1rn-/-Rag2-/-DO11.10 Tg IL-6 MHC ClassII F759 TCR (Murakami et al., 2011) IL-17 DO11.10 (OVA γδT Il1rn-/34 Il1rn-/- (unpublished observation) T 4-2; IL-1 Th17 IL-1R1 IL-23R Th17 IL-23R (McGeachy et al., 2009) IL-23 IL-7R et al., 2010) (Liu Th17 γδ+T IL-1R1 CD4+ unpublished observation IL-23R naïve T IL-23 IL-23 IL-1R IL-1R1 IL-1 IL-1R1 IL-1 IL-1 IL-1 Th17 Akt-mTOR MAPK NF-κB IL-17 NF-kB IL-17 c-Rel (Ruan et al., 2011) c-Rel Rel-A IL-17 Rel-A 35 IL-17 αβ+T IL-17 (Powolny-Budnicka et al., 2011) 4-3; IL-6 IL-1 Th17 IL-6 Th17 al., 2011) IL-1 IL-6 IL-1 IL-21 (Hu et Th17 IL-6 IL-1 Th17 (Figure 18b) IL-6 Th17 IL-6 Th17 (Manel et al., 2008) Th17 Th17 Il1rn-/- IL-23 IL-23 (Cho et al., 2006) IL-6 Th17 IL-23 Th17 4 4; STAT3 STAT3 IL-6/IL-21/IL-23 Th17 STAT3 Th17 STAT3 IL-21 IL-6 Th17 Th17 (figure 6) Th17 STAT 36 IL-21 2005) STAT5 STAT3 STAT1 5 STAT3 (Yang et al., 2011) L-6 (Leonard and Spolski, STAT3 IL-21 Th17 Th17 LIF(Leukemia inhibitory factor) IL-6 IL-27 gp130 STAT3 (Batten et al., 2006; Cao et al., 2011; Stumhofer et al., 2006) LIF STAT3 ERK IL-27 IL-27 IL-1 IL-1 SOCS3 STAT1 SOCS3 STAT1 STAT3 Th17 STAT3 Th17 IL-6 IL-1 Foxp3 IL-1 Th17 (Figure 20) 37 (Il1rn-/-) IL-1 IL-1 Th17 Il1rn-/- IL-6 Il1rn-/-Il6-/- Il1rn-/- IL-6 Il1rn-/-Il6-/- Th17 IL-6 Th17 IL-1 IL-6 naïve CD4+T Th17 Il6-/- TCR naïve T TGF-β IL-1+IL-21 Th17 TGF-β TGF-β Nfkbiz Batf IL-1 Th17 naïve T IL-1R1 Th17 Th17 IL-1 Th17 Th17 IL-21 Rag2-/- IL-1 IL-1 Th17 Foxp3 IL-6 IL-21 TGF+IL-21 TGF-β IL-1 IL-1 TGF-β IL-1 Th17 38 naïve T (Abbreviations) 7AAD; 7-Amino-ActinomycinD APC; Allophycocyanin Ahr; Aryl Hydrocarbon Receptor Batf; Basic leucine zipper transcription factor Blimp-1; B lymphocyte induced maturation protein 1 CFSE; Carboxyfluorescein succinimidyl ester CIA; collagen-induced arthritis CTLA-4; Cytotoxic T-Lymphocyte Antigen 4 EDTA; ethylenediaminetetraacetic acid FACS; Fluorescence-activated cell sorting FBS; Fetal Bovine Serum FITC; fluorescein isothiocyanate Foxp3; the forkhead box P3 GATA3; GATA-binding protein 3 GITR; Glucocorticoid-induced TNF-receptor HTLV; Human T-lymphotropic Virus Hif1a; Hypoxia-inducible factor 1, alpha subunit ICOS; inducible T-cell co-stimulator IL-1RI; type I IL-1R IL-1Ra; IL-1 receptor antagonist IL; Interleukin IRF4; Interferon regulatory factor 4 39 LIF; Leukemia inhibitory factor LPL; lamina proprial lymphocytes LPS; Lipopolysaccharide MAPK; mitogen-actiated protein kinase MCM; Macrophage conditioned media MFI; Mean Fluorescent Intensity MHC; major histocompatibility complex Myd88; myeloid differentiation factor 88 NF-kB; nuclear factor kappa-light-chain-enhancer of activated B cells Nfkbiz; NF-kappa-B inhibitor zeta OVA; ovalbumin PBS; Phosphate buffered saline PE; Phycoerythrin PI3K; Phosphoinositide 3-kinase PKCθ; Protein kinase C θ PMA; Phorbol 12-Myristate 13-acetate R; receptor RA; rheumatoid arthritis ROR-γt; retinoic acid receptor-related orphan receptor Rag2; recombination activating gene 2 Rorc; RORgamma SIGIRR; Single Ig IL-1-related receptor SPF; specific pathogen-free conditions 40 STAT; Signal Transduction and Activator of Transcription T-bet; T box expressed in T cells TCR; T cell receptor TGF; transforming growth factor Tg; transgenic iTreg; induced regulatory T cell mTOR; mammalian target of Rapamycin rpm; rotation per minute 41 Abdollahi-Roodsaz, S., L.A.B. Joosten, M.I. Koenders, I. Devesa, M.F. Roelofs, T. Radstake, M. Heuvelmans-Jacobs, S. Akira, M.J.H. Nicklin, F. Ribeiro-Dias, and W.B. Van den Berg. 2008. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. Journal of Clinical Investigation 118:205-216. Adachi, O., T. Kawai, K. Takeda, M. Matsumoto, H. Tsutsui, M. Sakagami, K. Nakanishi, and S. Akira. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143-150. Ahern, P.P., C. Schiering, S. Buonocore, M.J. McGeachy, D.J. Cua, K.J. Maloy, and F. Powrie. 2010. Interleukin-23 Drives Intestinal Inflammation through Direct Activity on T Cells. Immunity 33:279-288. Aksentijevich, I., S.L. Masters, P.J. Ferguson, P. Dancey, J. Frenkel, A. van Royen-Kerkhoff, R. Laxer, U. Tedgard, E.W. Cowen, T.-H. Pham, M. Booty, J.D. Estes, N.G. Sandler, N. Plass, D.L. Stone, M.L. Turner, S. Hill, J.A. Butman, R. Schneider, P. Babyn, H.I. El-Shanti, E. Pope, K. Barron, X. Bing, A. Laurence, C.-C.R. Lee, D. Chapelle, G.I. Clarke, K. Ohson, M. Nicholson, M. Gadina, B. Yang, B.D. Korman, P.K. Gregersen, P.M. van Hagen, A.E. Hak, M. Huizing, P. Rahman, D.C. Douek, E.F. Remmers, D.L. Kastner, and R. Goldbach-Mansky. 2009. An Autoinflammatory Disease with Deficiency of the Interleukin-1-Receptor Antagonist. New England Journal of Medicine 360:2426-2437. Alexander, M.R., C.W. Moehle, J.L. Johnson, Z. Yang, J.K. Lee, C.L. Jackson, and G.K. Owens. 2012. Genetic inactivation of IL-1 signaling enhances atherosclerotic plaque instability and reduces outward vessel remodeling in advanced atherosclerosis in mice. Journal of Clinical Investigation 122:70-79. Batten, M., J. Li, S. Yi, N.M. Kljavin, D.M. Danilenko, S. Lucas, J. Lee, F.J. de Sauvage, and N. Ghilardi. 2006. Interleukin 27 limits autoimmune encephalomyelitis by suppressing the development of interleukin 17-producing T cells. Nature Immunology 7:929-936. Bauge, C., F. Legendre, S. Leclercq, J.M. Efissalde, J.P. Pujol, P. Galera, and K. Boumediene. 2007. Interleukin-1 beta impairment of transforming growth factor beta 1 signaling by down-regulation of transforming growth factor 42 beta receptor type II and up-regulation of smad7 in human articular Chondrocytes. Arthritis and Rheumatism 56:3020-3032. Bettelli, E., Y.J. Carrier, W.D. Gao, T. Korn, T.B. Strom, M. Oukka, H.L. Weiner, and V.K. Kuchroo. 2006. Reciprocal developmental pathways for the generation of pathogenic effector T(H)17 and regulatory T cells. Nature 441:235-238. Bubier, J.A., T.J. Sproule, O. Foreman, R. Spolski, D.J. Shaffer, H.C. Morse, III, W.J. Leonard, and D.C. Roopenian. 2009. A critical role for IL-21 receptor signaling in the pathogenesis of systemic lupus erythematosus in BXSB-Yaa mice. Proceedings of the National Academy of Sciences of the United States of America 106:1518-1523. Cao, W., Y. Yang, Z. Wang, A. Liu, L. Fang, F. Wu, J. Hong, Y. Shi, S. Leung, C. Dong, and J.Z. Zhang. 2011. Leukemia Inhibitory Factor Inhibits T Helper 17 Cell Differentiation and Confers Treatment Effects of Neural Progenitor Cell Therapy in Autoimmune Disease. Immunity 35:273-284. Cho, M.L., J.W. Kang, Y.M. Moon, H.J. Nam, J.Y. Jhun, S.B. Heo, H.T. Jin, S.Y. Min, J.H. Ju, K.S. Park, Y.G. Cho, C.H. Yoon, S.H. Park, Y.C. Sung, and H.Y. Kim. 2006. STAT3 and NF-kappa B signal pathway is required for IL-23-mediated IL-17 production in spontaneous arthritis animal model IL-1 receptor antagonist-deficient mice. Journal of Immunology 176:5652-5661. Chung, Y., S.H. Chang, G.J. Martinez, X.O. Yang, R. Nurieva, H.S. Kang, L. Ma, S.S. Watowich, A.M. Jetten, Q. Tian, and C. Dong. 2009. Critical Regulation of Early Th17 Cell Differentiation by Interleukin-1 Signaling. Immunity 30:576-587. Cohen, S., E. Hurd, J. Cush, M. Schiff, M.E. Weinblatt, L.W. Moreland, J. Kremer, M.B. Bear, W.J. Rich, and D. McCabe. 2002. Treatment of rheumatoid arthritis with anakinra, a recombinant human interleukin-1 receptor antagonist, in twenty-four-week, combination with multicenter, methotrexate - Results randomized, of a double-blind, placebo-controlled trial. Arthritis and Rheumatism 46:614-624. Cretney, E., A. Xin, W. Shi, M. Minnich, F. Masson, M. Miasari, G.T. Belz, G.K. Smyth, M. Busslinger, S.L. Nutt, and A. Kallies. 2011. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nature Immunology 12:304-U353. 43 Dang, E.V., J. Barbi, H.-Y. Yang, D. Jinasena, H. Yu, Y. Zheng, Z. Bordman, J. Fu, Y. Kim, H.-R. Yen, W. Luo, K. Zeller, L. Shimoda, S.L. Topalian, G.L. Semenza, C.V. Dang, D.M. Pardoll, and F. Pan. 2011. Control of T(H)17/T(reg) Balance by Hypoxia-Inducible Factor 1. Cell 146:772-784. de Lafaille, M.A.C., and J.J. Lafaille. 2009. Natural and Adaptive Foxp3(+) Regulatory T Cells: More of the Same or a Division of Labor? Immunity 30:626-635. Delgoffe, G.M., K.N. Pollizzi, A.T. Waickman, E. Heikamp, D.J. Meyers, M.R. Horton, B. Xiao, P.F. Worley, and J.D. Powell. 2011. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nature Immunology 12:295-U117. Dinarello, C.A. 2009. Immunological and Inflammatory Functions of the Interleukin-1 Family. Annual Review of Immunology 27:519-550. Durant, L., W.T. Watford, H.L. Ramos, A. Laurence, G. Vahedi, L. Wei, H. Takahashi, H.W. Sun, Y. Kanno, F. Powrie, and J.J. O'Shea. 2010. Diverse Targets of the Transcription Factor STAT3 Contribute to T Cell Pathogenicity and Homeostasis. Immunity 32:605-615. Furst, D.E. 2004. Anakinra: Review of recombinant human interieukin-1 receptor antagonist in the treatment of rheumatoid arthritis. Clinical Therapeutics 26:1960-1975. Genovese, M.C., F. Van den Bosch, S.A. Roberson, S. Bojin, I.M. Biagini, P. Ryan, and J. Sloan-Lancaster. 2010. LY2439821, a Humanized Anti-Interleukin-17 Monoclonal Antibody, in the Treatment of Patients With Rheumatoid Arthritis A Phase I Randomized, Double-Blind, Placebo-Controlled, Proof-of-Concept Study. Arthritis and Rheumatism 62:929-939. Glaccum, M.B., K.L. Stocking, K. Charrier, J.L. Smith, C.R. Willis, C. Maliszewski, D.J. Livingston, J.J. Peschon, and P.J. Morrissey. 1997. Phenotypic and functional characterization of mice that lack the type I receptor for IL-1. Journal of Immunology 159:3364-3371. Gulen, M.F., Z.Z. Kang, K. Bulek, W. Youzhong, T.W. Kim, Y. Chen, C.Z. Altuntas, K.S. Bak-Jensen, M.J. McGeachy, J.S. Do, H. Xiao, G.M. Delgoffe, B.K. Min, J.D. Powell, V.K. Tuohy, D.J. Cua, and X.X. Li. 2010. The Receptor SIGIRR Suppresses Th17 Cell Proliferation via Inhibition of the 44 Interleukin-1 Receptor Pathway and mTOR Kinase Activation. Immunity 32:54-66. Guo, L.Y., G. Wei, J.F. Zhu, W. Liao, W.J. Leonard, K.J. Zhao, and W. Paul. 2009. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proceedings of the National Academy of Sciences of the United States of America 106:13463-13468. Hirahara, K., K. Ghoreschi, A. Laurence, X.-P. Yang, Y. Kanno, and J.J. O'Shea. 2010. Signal transduction pathways and transcriptional regulation in Th17 cell differentiation. Cytokine & Growth Factor Reviews 21:425-434. Horai, R., M. Asano, K. Sudo, H. Kanuka, M. Suzuki, M. Nishihara, M. Takahashi, and Y. Iwakura. 1998. Production of mice deficient in genes for interleukin (IL)-1 alpha, IL-1 beta, IL-1 alpha/beta, and IL-1 receptor antagonist shows that IL-1 beta is crucial in turpentine-induced fever development and glucocorticoid secretion. Journal of Experimental Medicine 187:1463-1475. Horai, R., A. Nakajima, K. Habiro, M. Kotani, S. Nakae, T. Matsuki, A. Nambu, S. Saijo, H. Kotaki, K. Sudo, A. Okahara, H. Tanioka, T. Ikuse, N. Ishii, P.L. Schwartzberg, R. Abe, and Y. Iwakura. 2004. TNF-alpha is crucial for the development of autoimmune arthritis in IL-1 receptor antagonist-deficient mice. Journal of Clinical Investigation 114:1603-1611. Horai, R., S. Saijo, M. Tanioka, S. Nakae, K. Sudo, A. Okahara, T. Ikuse, M. Asano, and Y. Iwakura. 2000. Development of chronic inflammatory arthropathy resembling rheumatoid arthritis in interleukin 1 receptor antagonist-deficient mice. Journal of Experimental Medicine 191:313-320. Hu, W., T.D. Troutman, R. Edukulla, and C. Pasare. 2011. Priming Microenvironments Dictate Cytokine Requirements for T Helper 17 Cell Lineage Commitment. Immunity 35:1010-1022. Hueber, W., D.D. Patel, T. Dryja, A.M. Wright, I. Koroleva, G. Bruin, C. Antoni, Z. Draelos, M.H. Gold, P. Durez, P.P. Tak, J.J. Gomez-Reino, C.S. Foster, R.Y. Kim, C.M. Samson, N.S. Falk, D.S. Chu, D. Callanan, N. Quan Dong, K. Rose, A. Haider, F. Di Padova, G. Psoriasis Study, G. Rheumatoid Arthrit Study, and G. Uveitis Study. 2010. Effects of AIN457, a Fully Human Antibody to Interleukin-17A, on Psoriasis, Rheumatoid Arthritis, and Uveitis. Science Translational Medicine 2: Huehn, J., J.K. Polansky, and A. Hamann. 2009. Epigenetic control of FOXP3 45 expression: the key to a stable regulatory T-cell lineage? Nature Reviews Immunology 9:83-89. Ichiyama, K., H. Yoshida, Y. Wakabayashi, T. Chinen, K. Saeki, M. Nakaya, G. Takaesu, S. Hori, A. Yoshimura, and T. Kobayashi. 2008. Foxp3 inhibits ROR gamma t-mediated IL-17A mRNA transcription through direct interaction with ROR gamma t. Journal of Biological Chemistry 283:17003-17008. Ishida, Y., T. Kondo, A. Kimura, K. Matsushima, and N. Mukaida. 2006. Absence of IL-1 receptor antagonist impaired wound healing along with aberrant NF-kappa B activation and a reciprocal suppression of TGF-beta signal pathway. Journal of Immunology 176:5598-5606. Ishihara, K., S. Sawa, H. Ikushima, S. Hirota, T. Atsumi, D. Kamimura, S.J. Park, M. Murakami, Y. Kitamura, Y. Iwakura, and T. Hirano. 2004. The point mutation of tyrosine 759 of the IL-6 family cytokine receptor gp130 synergizes with HTLV-1 pX in promoting rheumatoid arthritis-like arthritis. International Immunology 16:455-465. Ivanov, I.I., B.S. McKenzie, L. Zhou, C.E. Tadokoro, A. Lepelley, J.J. Lafaille, D.J. Cua, and D.R. Littman. 2006. The orphan nuclear receptor ROR gamma t directs the differentiation program of proinflammatory IL-17(+) T helper cells. Cell 126:1121-1133. Iwakura, Y., H. Ishigame, S. Saijo, and S. Nakae. 2011. Functional Specialization of Interleukin-17 Family Members. Immunity 34:149-162. Iwakura, Y., S. Nakae, S. Saijo, and H. Ishigame. 2008. The roles of IL-17A in inflammatory immune responses and host defense against pathogens. Immunological Reviews 226:57-79. Jones, G., A. Sebba, J. Gu, M.B. Lowenstein, A. Calvo, J.J. Gomez-Reino, D.A. Siri, M. Tomsic, E. Alecock, T. Woodworth, and M.C. Genovese. 2010. Comparison of tocilizumab monotherapy versus methotrexate monotherapy in patients with moderate to severe rheumatoid arthritis: the AMBITION study. Annals of the Rheumatic Diseases 69:88-96. Kimura, A., T. Naka, K. Nohara, Y. Fujii-Kuriyama, and T. Kishimoto. 2008. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proceedings of the National Academy of Sciences of the United States of America 105:9721-9726. Kopf, M., H. Baumann, G. Freer, M. Freudenberg, M. Lamers, T. Kishimoto, R. 46 Zinkernagel, H. Bluethmann, and G. Kohler. 1994. IMPAIRED IMMUNE AND ACUTE-PHASE RESPONSES IN INTERLEUKIN-6-DEFICIENT MICE. Nature 368:339-342. Korn, T., E. Bettelli, W. Gao, A. Awasthi, A. Jager, T.B. Strom, M. Oukka, and V.K. Kuchroo. 2007. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature 448:484-U489. Korn, T., E. Bettelli, M. Oukka, and V.K. Kuchroo. 2009. IL-17 and Th17 Cells. Annual Review of Immunology 27:485-517. Kryczek, I., S. Wei, L.H. Vatan, J. Escara-Wilke, W. Szeliga, E.T. Keller, and W. Zou. 2007. Cutting edge: Opposite effects of IL-1 and IL-2 on the regulation of IL-17(+) T cell pool IL-1 subverts IL-2-mediated suppression. Journal of Immunology 179:1423-1426. Lee, Y.K., H. Turner, C.L. Maynard, J.R. Oliver, D.Q. Chen, C.O. Elson, and C.T. Weaver. 2009. Late Developmental Plasticity in the T Helper 17 Lineage. Immunity 30:92-107. LeibundGut-Landmann, S., O. Gross, M.J. Robinson, F. Osorio, E.C. Slack, S.V. Tsoni, E. Schweighoffer, V. Tybulewicz, G.D. Brown, J. Ruland, and C.R.E. Sousa. 2007. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nature Immunology 8:630-638. Leonard, W.J., and R. Spolski. 2005. Interleukin-21: A modulator of lymphoid proliferation, apoptosis and differentiation. Nature Reviews Immunology 5:688-698. Li, T., and S.H. He. 2006. Induction of IL-6 release from human T cells by PAR-1 and PAR-2 agonists. Immunology and Cell Biology 84:461-466. Liu, X.B., S. Leung, C.X. Wang, Z. Tan, J. Wang, T.B. Guo, L. Fang, Y.G. Zhao, B. Wan, X. Qin, L.M. Lu, R.S. Li, H. Pan, M.J. Song, A.L. Liu, J. Hong, H.T. Lu, and J.Z. Zhang. 2010. Crucial role of interleukin-7 in T helper type 17 survival and expansion in autoimmune disease. Nature Medicine 16:191-U197. Manel, N., D. Unutmaz, and D.R. Littman. 2008. The differentiation of human T-H-17 cells requires transforming growth factor-beta and induction of the nuclear receptor ROR gamma t. Nature Immunology 9:641-649. McGeachy, M.J., Y. Chen, C.M. Tato, A. Laurence, B. Joyce-Shaikh, W.M. Blumenschein, T.K. McClanahan, J.J. O'Shea, and D.J. Cua. 2009. The 47 interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nature Immunology 10:314-324. McInnes, I.B., and G. Schett. 2007. Cytokines in the pathogenesis of rheumatoid arthritis. Nature Reviews Immunology 7:429-442. Monteleone, G., F. Pallone, and T.T. MacDonald. 2008. Interleukin-21: a critical regulator of the balance between effector and regulatory T-cell responses. Trends in Immunology 29:290-294. Murakami, M., Y. Okuyama, H. Ogura, S. Asano, Y. Arima, M. Tsuruoka, M. Harada, M. Kanamoto, Y. Sawa, Y. Iwakura, K. Takatsu, D. Kamimura, and T. Hirano. 2011. Local microbleeding facilitates IL-6- and IL-17-dependent arthritis in the absence of tissue antigen recognition by activated T cells. Journal of Experimental Medicine 208:103-114. Nakae, S., S. Saijo, R. Horai, K. Sudo, S. Mori, and Y. Iwakura. 2003. IL-17 production from activated T cells is required for the spontaneous development of destructive arthritis in mice deficient in IL-1 receptor antagonist. Proceedings of the National Academy of Sciences of the United States of America 100:5986-5990. Nakajima, A., T. Matsuki, M. Komine, A. Asahina, R. Horai, S. Nakae, H. Ishigame, S. Kakuta, S. Saijo, and Y. Iwakura. 2010. TNF, but Not IL-6 and IL-17, Is Crucial for the Development of T Cell-Independent Psoriasis-Like Dermatitis in Il1rn(-/-) Mice. Journal of Immunology 185:1887-1893. Nakayamada, S., Y. Kanno, H. Takahashi, D. Jankovic, K.T. Lu, T.A. Johnson, H.-w. Sun, G. Vahedi, O. Hakim, R. Handon, P.L. Schwartzberg, G.L. Hager, and J.J. O'Shea. 2011. Early Th1 Cell Differentiation Is Marked by a Tfh Cell-like Transition. Immunity 35:919-931. Nishimoto, N., N. Miyasaka, K. Yamamoto, S. Kawai, T. Takeuchi, and J. Azuma. 2009. Long-term safety and efficacy of tocilizumab, an anti-IL-6 receptor monoclonal antibody, in monotherapy, in patients with rheumatoid arthritis (the STREAM study): evidence of safety and efficacy in a 5-year extension study. Annals of the Rheumatic Diseases 68:1580-1584. Okamoto, K., Y. Iwai, M. Oh-hora, M. Yamamoto, T. Morio, K. Aoki, K. Ohya, A.M. Jetten, S. Akira, T. Muta, and H. Takayanagi. 2010. I kappa B zeta regulates T(H)17 development by cooperating with ROR nuclear receptors. 48 Nature 464:1381-U1313. Powolny-Budnicka, I., M. Riemann, S. Taenzer, R.M. Schmid, T. Hehlgans, and F. Weih. 2011. ReIA and ReIB Transcription Factors in Distinct Thymocyte Populations Control Lymphotoxin-Dependent Interleukin-17 Production in gamma delta T Cells. Immunity 34:364-374. Puga, I., M. Cols, C.M. Barra, B. He, L. Cassis, M. Gentile, L. Comerma, A. Chorny, M. Shan, W. Xu, G. Magri, D.M. Knowles, W. Tam, A. Chiu, J.B. Bussel, S. Serrano, J. Antonio Lorente, B. Bellosillo, J. Lloreta, N. Juanpere, F. Alameda, T. Baro, R. Diaz de Heredia, N. Toran, A. Catala, M. Torrebadell, C. Fortuny, V. Cusi, C. Carreras, G.A. Diaz, J.M. Blander, C.-M. Farber, G. Silvestri, C. Cunningham-Rundles, M. Calvillo, C. Dufour, L.D. Notarangelo, V. Lougaris, A. Plebani, J.-L. Casanova, S.C. Ganal, A. Diefenbach, J. Ignacio Arostegui, M. Juan, J. Yaguee, N. Mahlaoui, J. Donadieu, K. Chen, and A. Cerutti. 2012. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nature Immunology 13:170-180. Ramonda, R., A. Lo Nigro, V. Modesti, L. Nalotto, E. Musacchio, L. Iaccarino, L. Punzi, and A. Doria. 2011. Atherosclerosis in psoriatic arthritis. Autoimmunity Reviews 10:773-778. Reddy, S., S. Jia, R. Geoffrey, R. Lorier, M. Suchi, U. Broeckel, M.J. Hessner, and J. Verbsky. 2009. BRIEF REPORT An Autoinflammatory Disease Due to Homozygous Deletion of the IL1RN Locus. New England Journal of Medicine 360:2438-2444. Ruan, Q., V. Kameswaran, Y. Zhang, S. Zheng, J. Sun, J. Wang, J. DeVirgiliis, H.-C. Liou, A.A. Beg, and Y.H. Chen. 2011. The Th17 immune response is controlled by the Rel-ROR gamma-ROR gamma T transcriptional axis. Journal of Experimental Medicine 208:2321-2333. Schmitt, N., R. Morita, L. Bourdery, S.E. Bentebibel, S.M. Zurawski, J. Banchereau, and H. Ueno. 2009. Human Dendritic Cells Induce the Differentiation of Interleukin-21-Producing T Follicular Helper-like Cells through Interleukin-12. Immunity 31:158-169. Shinkai, Y., G. Rathbun, K.P. Lam, E.M. Oltz, V. Stewart, M. Mendelsohn, J. Charron, M. Datta, F. Young, A.M. Stall, and F.W. Alt. 1992. RAG-2-DEFICIENT MICE LACK MATURE LYMPHOCYTES OWING TO INABILITY TO INITIATE V(D)J REARRANGEMENT. Cell 68:855-867. 49 Sims, J.E., and D.E. Smith. 2010. The IL-1 family: regulators of immunity. Nature Reviews Immunology 10:89-102. Spolski, R., and W.J. Leonard. 2010. IL-21 and T follicular helper cells. International Immunology 22:7-12. Stritesky, G.L., N. Yeh, and M.H. Kaplan. 2008. IL-23 Promotes Maintenance but Not Commitment to the Th17 Lineage. Journal of Immunology 181:5948-5955. Stumhofer, J.S., A. Laurence, E.H. Wilson, E. Huang, C.M. Tato, L.M. Johnson, A.V. Villarino, Q.L. Huang, A. Yoshimura, D. Sehy, C.J.M. Saris, J.J. O'Shea, L. Hennighausen, M. Ernst, and C.A. Hunter. 2006. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nature Immunology 7:937-945. Sutton, C., C. Brereton, B. Keogh, K.H.G. Mills, and E.C. Lavelle. 2006. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. Journal of Experimental Medicine 203:1685-1691. Tak, P.P., and J.R. Kalden. 2011. Advances in rheumatology: new targeted therapeutics. Arthritis Research & Therapy 13: Van Kooten, C., I. Rensink, D. Pascual-Salcedo, R. Van Oers, and L. Aarden. 1991. MONOKINE PRODUCTION BY HUMAN T CELLS IL-1-ALPHA PRODUCTION RESTRICTED TO MEMORY T CELLS. Journal of Immunology 146:2654-2658. Veldhoen, M., R.J. Hocking, R.A. Flavell, and B. Stockinger. 2006. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nature Immunology 7:1151-1156. Wei, L., A. Laurence, K.M. Elias, and J.J. O'Shea. 2007. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. Journal of Biological Chemistry 282:34605-34610. Yang, X.-P., K. Ghoreschi, S.M. Steward-Tharp, J. Rodriguez-Canales, J. Zhu, J.R. Grainger, K. Hirahara, H.-W. Sun, L. Wei, G. Vahedi, Y. Kanno, J.J. O'Shea, and A. Laurence. 2011. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nature Immunology 12:247-U284. 50 Yang, X.O., R. Nurieva, G.J. Martinez, H.S. Kang, Y. Chung, B.P. Pappu, B. Shah, S.H. Chang, K.S. Schluns, S.S. Watowich, X.-H. Feng, A.M. Jetten, and C. Dong. 2008a. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29:44-56. Yang, X.O., R. Nurieva, G.J. Martinez, H.S. Kang, Y. Chung, B.P. Pappu, B. Shah, S.H. Chang, K.S. Schluns, S.S. Watowich, X.H. Feng, A.M. Jetten, and C. Dong. 2008b. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29:44-56. Zhou, L., Ivanov, II, R. Spolski, R. Min, K. Shenderov, T. Egawa, D.E. Levy, W.J. Leonard, and D.R. Littman. 2007. IL-6 programs TH-17 cell differentiation by promoting the sequential engagement of the IL-21 and IL-23 pathways. In 176. Zhou, L., and D.R. Littman. 2009. Transcriptional regulatory networks in Th17 cell differentiation. Current Opinion in Immunology 21:146-152. Zhou, L., J.E. Lopes, M.M.W. Chong, Ivanov, II, R. Min, G.D. Victora, Y.L. Shen, J.G. Du, Y.P. Rubtsov, A.Y. Rudensky, S.F. Ziegler, and D.R. Littman. 2008. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing ROR gamma t function. Nature 453:236-U214. Zhou, X.Y., S.L. Bailey-Bucktrout, L.T. Jeker, C. Penaranda, M. Martinez-Llordella, M. Ashby, M. Nakayama, W. Rosenthal, and J.A. Bluestone. 2009. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nature Immunology 10:1000-U1104. 51 a b IL#6R & Human&IL#1Ra& TNF#α Figure 0-1 a, IL-6-TNF b, IL-6 IL-1 TNF Human IL-1Ra ( IL-6R ) TNF & a IL-1α IL-1R1 NFkB AP-1 IL-1R AcP IL-1β IL-1Ra Cell membrane Il1rn-/- Il1rn+/+ c 100! Incidence of RA (%)! b Il1rn-/-Il17+/+! 50! Il1rn-/-Il17-/-! 0! 0! 5! 10! 15! Age (week)! Horai et al., J. Exp. Med.,2000! d Figure 0-2 IL-1Ra a, IL-1Ra b, IL-1Ra c, d, IL-1Ra IL-17 T IL-1 IL-17 T nude Nakae et al., ! Proc. Natl. Acad. Sci. 2003 20! β Figure 0-3 Th17 T IL-17 CD4+ CD4+ T TGF-β IL-6 T IL-17 T Th17 Th17 TCR# TGF-β# IL-6/ IL-21! IL-23 IL-1 IFN-γ# IL-2 IL-27! IL-4! SOCS3 STAT3 Foxp3 STAT5 STAT1 STAT6 Irf4 AHR Rorc FoxP3 Ahr RORγt Rora NFATc1 BATF RUNX1 RORγt STAT3 STAT5 RORα Nfkbiz IkBζ Il17a!Promoter!&! Enhancer Il17a Figure 0-4 Th17 IL-17 Th17 Th17 RORγt Th17 RORγt STAT3 Th17 Th17 Foxp3 STAT3 Figure 0-5 Th17 Treg Th17 Th17 TGF-β Foxp3 RORγ IL-6 RORγt Foxp3 RORγ STAT3 Foxp3 TGF-β RORγ Foxp3+ Th17 a 6 100 * 5 Severity (Score) Incidence (%) 80 60 40 Il1rn-/20 Il1rn-/-Il6-/- b Wt 12 week 4 20 Il6-/- Il1rn-/- 0.18& 1.3& 8 12 16 20 week Il1rn-/-Il6-/0.99& IL-17 0.16& 16 2 0 0 8 3 1 Il1rn-/-Il6+/- 4 4 CD4 Figure 1. Il1rn-/-Il6-/(a) : Il1rn-/: Il6-/-, (b WT, in vitro (%) χ2 Il1rn-/-, PMA/ionomycin CD4+ Th17 n=15-22/ *, P < 0.05: Il1rn-/- Il1rn-/-Il6-/- Il6-/-Il1rn-/IL-17 LNs CD8+ cells ! (106 cells) 0" 4" * Il1rn-/80" * 5" 0" 0" * 3" 2" 1" 0" Il6-/25" * * 60" * * 50" 40" 30" 20" 10" 0" * 40" 20" * 20" 15" 10" 5" 0" 3-4 70" 60" 50" 40" 20" 12" 10" 8" 6" 4" 2" 0" Figure 2. Il1rn-/-Il6-/- ± SD *P < 0.05. 2 B220+ cells ! (%) 30" 20" 10" Spleen cells (106 cells) 150" 100" 50" Spleen/body weight (µg) 200" * 15" 10" 5" * CD4+ cells ! (%) 0" Spleen 5" B220+ cells ! (106 cells) 10" Spleen 5" Spleen 15" CD4+ cells ! (106 cells) 10" 60" Spleen 20" 250" CD8+ cells ! (%) 10" * p=0.09 Spleen 5" * * CD8+ cells ! (106 cells) 15" * LNs B220+ cells ! (%) LNs cells (106 cells) 25" Spleen 15" * LNs CD4+ cells ! (%) LNs B220+ cells ! (106 cells) 30" LNs CD8+ cells ! (%) LNs CD4+ cells ! (106 cells) 35" [LNs] [Spleen] 300" 12" 10" 8" 6" 4" 0" 2" 80" 0" 140" 120" 100" 0" 25" 0" 80" 60" 40" 20" 0" 50" 40" 30" 20" 10" 0" 20" 15" 10" 5" 0" * * LN IFNg+ (TCRb+CD4+) cells! LN Foxp3+ (TCRb+CD4+) cells! 2.5" * * LN IL-17+ (TCRb+CD4+) cells! n.s 2" 0.09" 0.08" 1.5" 1" 1.5" 1" 0.07" (106 cells) (106 cells) (106 cells) 2" * 0.06" 0.05" 0.04" 0.03" 0.5" 0.5" 0.02" 0.01" 0" 0" Figure 3. 0" Th T WT, Il6-/-, arthritic Il1rn-/-, Il6-/- , arthritic Il1rn-/-Il6-/PMA/ionomycin IFN-γ, IL-17A, Foxp3 3-4 ± SD 2 CD4+ Wt 0.6 2.7 Il6-/- medium 2.3 6.9 6.5 9.4 IL-1 1.6 IL-17 3.5 IFN-γ# Figure 4. IL-1 IL-6 CD4+ Th17 $/$ + WT Il6 & CD4 IL-1 anti-CD3 (4 µg/ ml), anti-CD28 (1µg/ml), anti-IL-4 (10 µg/ml), anti-IFN-γ (10 µg/ml) 48well Flat Bottom plate 5 5 PMA/ionomycine 5 FACS CD4+ (%) 5 Il21 4 LNs * * * * 3 2 1 0 3 Spleen * * 2.5 Il21 2 1.5 1 0.5 0 None stimulation PMA/Ionomycin Figure 5. WT, Il1rn-/-, Il6-/-, Il1rn-/-Il6-/mRNA real-time RT-PCR IL-21 PMA/ionomycin GAPDH mRNA n=3 2 Il21 mRNA 4 5 0.1 IL-1 1.2 0.86 TGF/IL-6 17 1.01 TGF/IL-1,6 33 0.07 IL-17 TGF 0.03 TGF/IL-1 c 10 0.9 1.2 0.11 TGF/IL-21 8.2 0.11 0 TGF/IL-1,21 18 100 0.11 Rorc medium Foxp3 a 0.06 50 0 1.5 medium IL-1 TGF TGF/IL-1 Il17a IFN-γ# b 5 1 0.5 0 TGF/IL-6 TGF/IL-1,6 TGF/IL-21 TGF/IL-1,21 Il1r1 6 4 2 Il21 0 0.3 IL-1R1" 0.2 Il22 0.1 0 14 12 10 8 6 4 2 0 Figure 6. IL-1 IL-6 Th17 (a-b) In vitro Th17 BALB/cA FACS naïve + CD4 T anti-CD3 (4 µg/ml) anti-CD28 (1 µg/ml) 10 ng/ml; anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml (TGF-β, 3 ng/ml; IL-6, 40 ng/ml; IL-21, 100 ng/ml; IL-1, 10 ng/ ml) IL-17A FN-γ IL-1R1 PMA-ionomycin FACS (%) (b) IL-1R a Anti-IL-1R1: ( ): (c) BALB/cA FACS naïve CD4+T anti-CD3 (4 µg/ml) anti-CD28 (1 µg/ml) 10 ng/ml; anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml 5 mRNA mRNA PCR GAPDH RNA 0.04 IL-17 1.9 Foxp3 0.02 0.04 1.5 0.07 3.2 0.11 2.9 5.0 IL-1α,β 0.1-100ng/ml Figure 7 naïve T IL-1 FACS naïve CD4+T anti-CD3 (4 µg/ml) ml; anti-IL-4, 10 µg/ml IL-1(0.1-100 ng/ml) 4 Foxp3 IL-17 anti-CD28 (1 µg/ml) anti-IFN-γ, 10 µg/ 4 PMA/ionomycin 2 IL-1R1 TGF-β TGF-β / IL-21 (20-1000ng/ml) Figure 8 IL-21 IL-1R1 FACS naïve CD4+T anti-CD3 (4 µg/ml) anti-CD28 (1 µg/ml) γ, 10 µg/ml; anti-IL-4, 10 µg/ml TGF-β (3 ng/ml) ; IL-21 (20, 100, 400, 1000 ng/ml) 4 PMA/ionomycin IL-1R1 TGF-β anti-IFN- 2 1st round TGF/IL-21 2nd round 4.8" 9.9" 3nd round 3.1" TGF 0.27" IL-17 5.53" 1.3" IFN-γ# 9.7" 3.5" TGF/ IL-21 6.0" 0.24" 17.3" 11.5" TGF/ IL-1,21 2.2" 1.34" 17.2" 12.2" TGF/ IL-1 1.9" 0.24" 2.2" 1.4" IL-1 0.79" 0.22" 0.83" 0.02" med 3.9" IL-17 3.9" IFN-γ# Figure 9 IL-1 TGF-β IL-17 FACS naïve CD4+T Th17 anti-CD3 (4 µg/ml) antiCD28 (1 µg/ml) 10 ng/ml; anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml; TGF-β, 3 ng/ml; IL-21, 100 ng/ ml 5 (1st round culture) anti-IFN-γ, anti-IL-4 anti-CD3 anti-CD28 PMA/ionomycin IL-17A IFN-γ FACS 3 3 d c Il17a Il21 Il22 medium 2 0 0.3 1 24.4 14.4 7.9 24.9 IL-21 0.2 10 Foxp3 Nfkbiz Batf Rorc 1.6 0 1 Gata3 2 0.1 1.2 IL-1 0 6 0.4 4 Rorc 0.8 0.01 0.1 1 IL-1 (ng/ml) 2 10 12 Foxp3 0 13.4 0.5 IL-23 0 3.4 8 4 0 0 21.2 11.6 Foxp3 MyD88+/- e 10 8 6 4 2 0 MyD88-/5 * Foxp3+ cells (%) 0.01 0.1 1 IL-1 (ng/ml) Tbx21 0 relative expression (fold) +TGF 0.1 0 b no TGF Il17a relative expression (fold) 4 Foxp3+ cells (%) a 4 3 2 1 0 Figure 10. IL-1 in vitro Foxp3 (a), IL-1 Th17 Il6-/FACS naïve CD4+T Th17 anti-CD3 (4 µg/ml) antiCD28 (1 µg/ml) anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml; TGF-β, 3 ng/ml; IL-21, 100 ng/ml 4 IL-1 (0-10 ng/ml) anti-CD3, anti-CD28 24 mRNA Real time PCR mRNA (a)& IL#17A IL-22 IL-21 IL-1 (b) Nfkbiz Batf Rorc Foxp3 GAPDH RNA (c-d), FACS naïve CD4+T Th17 anti-CD3 (4 µg/ml) anti-CD28 (1 µg/ml) anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml; TGF-β, 3 ng/ml; IL-21, 100 ng/ml 5 anti-CD3 anti-CD28 48 (c)Il17a Tbx21 Gata3 Rorc Foxp3 mRNA GAPDH RNA (d), PMA/ionomycin Foxp3 FACS ; TGF-β ; TGF-β (%) (e) MyD88+/MyD88-/FACS + naïve CD4 T Th17 anti-CD3 (4 µg/ml) anti-CD28 (1 µg/ml) anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml; TGF-β, 3 ng/ml; IL-21, 100 ng/ml 4 anti-CD3 anti-CD28 48 PMA/ionomycin Foxp3 FACS *P&<&0.05&(student&t#test) IL-17+ Foxp3+ Il1r1-/Wt 2.7 42 54 IL-17 Foxp3 Figure 11. IL-1R IL-17-Foxp3- IL-1R1 Th17 IL-17+ Foxp3+ FACS Il1r1-/- naïve CD4+T Th17 antiCD3 (4 µg/ml) anti-CD28 (1 µg/ml) anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml; TGF-β, 3 ng/ml; IL-21, 100 ng/ml 4 (1st round culture) 4 PMA/ionomycin IL-1R1 Foxp3 IL-17A WT T -/IL-1R1 Il1r1 T 2 71 * 2000 TGF 1600 59 36 TGF/IL-21 3.7 67 26 Foxp3 MFI (in Foxp3+ cell) 3.7 87 RORγt TGF/IL-6 CFSE 6.1 Foxp3 Foxp3CFSE 800 400 0 5.7 Foxp3+ 1200 TGF/IL-1,21 5.9 * * c Foxp3 b 24 TGF" Foxp3+ cell (%) in each division a 90" TGF/IL-21" 80" TGF/IL-1,21" 70" TGF/IL-6" 60" 50" 40" 30" * 20" 10" 0" 0" 1" 2" 3" 4" cell division 5" 6" Figure 12. IL-1 Foxp3 -/(a) Il6 FACS naïve CD4+T CFSE anti-CD3 (4 µg/ ml) anti-CD28 (1 µg/ml) anti-IFN-γ 10 µg/ml anti-IL-4 10 µg/ml 4 RORγt Foxp3 (%) (b) Foxp3 (Mean fluorescence intensity; MIF) + (a) (c) Foxp3 CFSE *P < 0.05 (student t-test) 3 b d 0.53 TGF 27 3.5 Foxp3+ cell population (%) " a TGF" 40& TGF+IL-1" medium 30& 20& IL#1 10& 0& TGF/IL-21 IL-21R-Fc " TGF" TGF+IL-1" IIL#21R#Fc 20000& 9.7 15000& 10000& 5.8 IL#1& IL#21R#Fc 5000& 0& pSTAT3 IL-21R-Fc " c e 10 Il6-/- Il1rn-/-Il6-/- Il1ab-/- 20 15 10 0 IL-1 (0.01-10 ng/ml) IL-21R-Fc 20 µg/ml IL-1 10 ng/ml 5 IL-1 0 ng/ml Foxp3+ cell (%) 25 Foxp3+ cells (%) IL-17 TGF/IL-21 IL-1 10ng Foxp3 Foxp3 MFI in Foxp3+ cells " 13 8 6 4 2 0 med TGF TGF/IL-1 Foxp3 IL-6 IL-21 (a) Il6-/FACS anti-CD3 (4 µg/ml) anti-CD28 (1 µg/ml) anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml; 4 PMA/ionomycin Foxp3 IL-17 (%) 2 (b-c) -/+ Il6 FACS naïve CD4 T Th17 anti-CD3 (4 µg/ml) antiCD28 (1 µg/ml) anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml; TGF-β, 3 ng/ml; IL-21, 100 ng/ml 4 IL-21R-Fc (1-15 µg/ml) anti-CD3 anti-CD28 2 Foxp3 + FACS : Foxp3 , : Foxp3 MFI (c-d) Th17 IL-1 (10 ng/ml) IL-21R-Fc (20 µg/ml) anti-CD3 anti-CD28 Foxp3 pSTAT3 *P<0.05 (student t-test)(e) IL-1 Foxp3 IL-6 IL-1α/IL-1β, -/-/-/-/IL-1Ra Il6 , Il1rn Il6 , Il1αβ FACS naïve CD4+T Th17 anti-CD3 (4 µg/ml) anti-CD28 (1 µg/ml) 10 ng/ml; anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ ml; TGF-β, 3 ng/ml; IL-21, 100 ng/ml 4 anti-CD3 anti-CD28 Foxp3 2 Figure 13. IL-1 naïve CD4+T Wt naïve T 1.2 Il1ab-/- naïve T 37 40 0.8 0.36 med 13 1.7 0.27 6.9 2.4 IL-6 0.8ng 11 IL-6 4ng 0.13 TGF/IL1 TGF 2.6 21 IL-6 20ng 27 IL-6 100ng TGF/IL1 TGF 9.8 14 2.3 IL-6 0.8ng 2.7 4.2 IL-6 4ng + TGF Foxp3" 34 0.37 IL-21 20ng 0.4 0.31 med 1.5 39 45 1.8 18 IL-6 100ng 13 IL-6 20ng + TGF 21 16 22 2.4 IL-21 100ng 2.9 IL-21 400ng + TGF 3.9 IL-21 1000ng 36 18 0.54 IL-21 20ng 23 17 1.7 IL-21 100ng 2.8 IL-21 400ng + TGF 2.7 IL-21 1000ng IL-17A" Figure 14. IL-6 IL-21 Foxp3 IL-17 IL-1 -/+ FACS Il1ab naïve CD4 T TGF-β: 3 ng/ml, IL-6: 40 ng/ml or IL-21: 100 ng/ml, anti-IFN-γ: 10 µg/ml, anti-IL-4: 10 µg/ml; anti-CD3 (4 µg/ml); anti-CD28 (1 µg/ml) 4 PMA/ionomycin IL-17A, Foxp3 (%) 4.5" 4" 3.5" 3" 2.5" 2" 1.5" 1" 0.5" 0" 2" 1.8" 1.6" 1.4" 1.2" 1" 0.8" 0.6" 0.4" 0.2" 0" Nfkbiz/Gapdh 2" 1.5" 1" 1.5" 1" 0.5" 0" 2" 1.8" 1.6" 1.4" 1.2" 1" 0.8" 0.6" 0.4" 0.2" 0" 2.5" 2" Il21/Gapdh Il17a/Gapdh 0.5" 2" Il22/Gapdh Foxp3/Gapdh 2.5" 2.5" Rorc/Gapdh 0" 2" 1.8" 1.6" 1.4" 1.2" 1" 0.8" 0.6" 0.4" 0.2" 0" Batf/Gapdh 3" 1.5" 1" 0.5" 0" 1.8" 1.6" IL#1 IL#21 Rorc Il21 1.4" & 1" 0.8" IL-21 Il1r1 1.2" IL#21 & Foxp3 Ba4 5 Il17 Il22 IL#1 0.6" IL#1 IL$1 IL#21 IL$21 N9biz 5 0.4" 0.2" 0" med" IL-1" Fc" Fc IL-1" IL-1 Figure 15. IL-1 IL-21 Il6-/FACS naïve CD4+T Th17 anti-CD3 (4 µg/ml) (1 µg/ml) anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml; TGF-β, 3 ng/ml; IL-21, 100 ng/ml 4 IL-1 (10 ng/ml) IL-21R-Fc (20 µg/ml) anti-CD3, anti-CD28 24 mRNA PCR mRNA anti-CD28 Real time LPL& LPL!Foxp3+!cell! (gated!on!CD4+TCR9b+) LPL!IFN9g+!! (gated!on!CD4+TCR9b+)! 35& 25& LPL!IL917+!%!! (gated!on!CD4+TCR9b+)! 14& 25& 10& 12& 20& 10& (%) 15& (%) (%)! 20& 15& 5& 0& 0& ① ② ③ ④ 8& 6& 10& 5& * 16& 30& 4& 2& ① ② ③ 0& ④ ① ② ③ Wild type" Il1rn-/-! Il6-/-! Il1rn-/-Il6-/- Figure 16. IL-1 IL-21 Il1rn-/- Il6-/- Il1rn-/-Il6-/Foxp3 IFN-γ LPL) IL-17 ) LPL FACS PMA/ionomycin 5 ④ a b * 130" Body weight (%) 120" 110" R1" 90" 17" 80" 70" 0" 1" 2" 3" 4" weeks after transfer 6" W" Il1r1-/-" 17" * c 12& MLN!IL917+!cell!(%)!! 15& 10& 5& 0& 10& 1.5& 8& 6& 4& W 0& Il1r1-/- Il17a-/- 0& LPL!IL917+!cell!(%)!! 10& 5& Il1r1-/- Il17a-/- W 20& 5& 4& 3& 2& 1& 0& 0& W Il1r1-/- Figure 17. IL-1 in vivo Il17a-/- Il1r1-/(a) test) (b) 6 (c) 6 MLN Il17a-/- Foxp3 IFN-γ 15& 10& 5& 0& W Il1r1-/- Il17a-/- Th17 IL-17 CD4+CD45RBhiCD25Rag2-/1 :Il1r1-/LPL) IL-17 W Il1r1-/- Il17a-/- * 6& 15& 1& 0.5& 2& 20& LPL!IFN9g+!cell!(%)!! MLN!Foxp3+!cell!(%)!! 20& 2& LPL!Foxp3+!cell!(%)!! MLN!IFN9g+!cell!(%)!! 25& test) 5" cm W" 100" * 10" 9" 8" 7" 6" 5" 4" 3" 2" 1" 0" FACS W Il1r1-/- Il17a-/- :Il1r1-/- p<0.05 (student t-test) ) PMA/ionomycin :Il1r1-/- p<0.05 (student t- 5 p<0.05 (student t- medium a LPS 5.9 zymosan 7.4 3.5 Wt MCM 3.4 3.7 4.8 6.8 0.4 2.8 Il6-/MCM 3.9 0.19 200012.2.800 Workspace.jo 1.1 Layout 16 11 Il1rn-/-Il6-/MCM 2.5 3.9 2.5 IL-17 7.9 IFN-γ# TGF b 10 2 0 0 1.25 10 4 47.7 10 5 10 10 3 10 2 0 Foxp3 10 5 10 2 10 3 IL-17 10 4 29 0 10 5 1.13 10 5 Foxp3 10 2 10 2 0 10 2 3.12 10 3 IL-17 10 4 10 5 10 3 IL-17 10 4 26.7 10 4 21.1 10 5 0.626 10 3 10 5 69.1 0 0.749 10 5 10 2 9.22 10 3 IL-17 10 4 16.9 10 5 0.822 10 4 10 3 10 3 10 2 IL-6 64.9 Th1767.4 0 TGC anti-CD3 anti-CD3 anti-CD28 PMA/ionomycin 27.9 10 3 IL-17 4 0 9.07 10 2 0 10 2 10 2 55.8 10 4 10 3 66.8 0 10 0 IL-17 Foxp3 10 4 10 3 10 5 0.831 10 3 24.6 10 3 IL-17 34 10 5 0.235 Figure 18. Il1rn-/67.6 -/-/-I (a) Il6 Il1rn l6-/(MCM) Il6-/-CD4+ 5 PMA/ionomycin (b) naïve T 5 10 2 4 0 1.75 10 4 Il1rn-/-Il6-/MCM 71.1 10 2 50.3 4.46 4 10 2 0 4 0 10 10 5 0.278 10 5 Foxp3 10 3 IL-17 Foxp3 Foxp3 10 10 2 0.257 10 3 10 2 46.3 3.97 4 Foxp3 10 3 10 5 MCM 10 5 10 0 Il6-/Il6-/MCM 0.203 Foxp3 Wt Wt MCM MCM Foxp3 10 52.2 4 Foxp3 10 5 TGF zymosan Zym MCM TGF LPS LPL MCM medium 0 10 2 10 3 IL-17 10 4 anti-CD28 anti-IFN-γ 0 7.64 10 5 10 2 14.8 10 3 IL-17 10 4 10 5 LPS zymosan anti-IFN-γ atni-IL-4 FACS atni-IL-4 0 TGF-β (3 ng/ml) CM FACS 5 5 12000" 25" * 15" 10" 5" 0" * * MFI (Foxp3+ cell) Foxp3+ cell (%) 20" * 10000" 8000" 6000" 4000" 2000" 0" Figure 19. IL-1 mTOR Foxp3 -/Il6 FACS naïve CD4+T Th17 anti-CD3 (4 µg/ml) anti-CD28 (1 µg/ml) anti-IFN-γ, 10 µg/ml; anti-IL-4, 10 µg/ml; TGF-β, 3 ng/ml; IL-21, 100 ng/ml 4 Rapamycin 50 ng/ml anti-CD3 anti-CD28 2 + Foxp3 FACS : Foxp3 , : Foxp3 MFI *P<0.05 (student t-test) !IL91 IL-21! IL-6 IL-21! IL-6 Th17 IL-1 IL-1 TGF NFκB" MAPK" mTOR" RORγ IL-1R1 Batf! Ikbiz STAT3 ! STAT3 Foxp3 naïve T IL-21R IL-21R IL-17 IL-22 IL-1R Foxp3+ Foxp3 IL#1 IL#1 IL#21 TGF +IL-21 Foxp3+ RORγt+ naïve T IL#1 Foxp3RORγt+ IL-17++ IL#1 Foxp3+ RORγt+ Foxp3RORγt+ IL-17++ Nfkbiz↑ Batf↑ TGF Foxp3RORγt+ IL-17++ Foxp3 downregulation Th17 cells Foxp3+ RORγt- iTreg Figure 20 Th17 Foxp3 RORγt IL-1 IL-21 IL-1R1 IL-1,IL-6,IL-21 naïve T Foxp3 TGF-β Th17 52