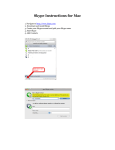
Download Observing with GISMO
Transcript
Observing with GISMO General: Observing with GISMO is little different from observing with IMACS using standard multi-slit masks. Hence much of the information in this cookbook is a repeat of the IMACS multi-slit cookbook. Please send any suggested clarifications, notes or other improvements to Mike Gladders at [email protected] Note: The software used at the telescope for aligning IMACS masks requires the existence of special files for each mask (referred to in this cookbook as ".align" and ".sub" files). This cookbook describes a method which uses only IRAF. Notation: multiple individual GISMO masks - (up to 4, plus a imaging 'open' mask) can be cut on a single piece of metal. We'll use the term 'Plate' to correspond the metal plate that contains multiple masks in GISMO, where 'mask' is a single setup for a single field, corresponding in a logical and execution sense to a normal IMACS mask. Afternoon: Make sure that the Instrument Specialist has loaded the plate that you will use that night onto GISMO, and loaded GISMO into IMACS, and powered up and initialized the instrument. The appendix provides details about how to do this. Remember that observers are required to submit mask files at least 6 weeks before the date of observation in order to insure that the masks will be ready for their run. If the IMACS GUI has not already been started by the Instrument Specialist, open an xterm, type "imacs ", and follow the instructions in the "Getting Started" section of the IMACS Manual. Take an image of each of the multi-slit masks. To do this, first ask the Instrument Specialist to open the telescope mirror covers. Start the Flat Field Screen gui by typing "ffs " in an xterm. Once the gui appears, click the mouse on the "IN" button to put the screen into the telescope beam. Ambient light in the dome should be sufficient to measure the slit positions. Now, in the IMACS MechGUI, make sure that the Hatch is in the "open" position, the calibration lamps are all turned off, and the CF-Guider is in the "out" position, and GISMO is inserted (IMPORTANT: See 'Setting Up GISMO' for details on inserting GISMO into the beam). The GISMO Mask must be set to the position with the mask that you intend to use, and Disperser should be set to "f/4-Imaging". f/4 is used for mask alignment, regardless of which camera you plan to use for spectroscopy! Finally, set the Filter to whatever blocking filter you intend to use. Now, in the IMACS CamGUI, set ExpTime to "10", Loops to "1", and ExpType to "Object". Finally, set Binning X and Binning Y to the 2x2, and then click on the "Start" button to take an exposure. Open an IRAF xgterm and the ds9 image display tool, and type "imacs" at the CL prompt to load the imacs package. Set up the input and output image names (the input is the image you just took) using "epar degismo".) Name the output image NAMEmask, e.g., if your mask is called S1028, call the output S1028mask. Run degismo on the image to produce an image of the “normal” IMACS focal plane; this task effectively undoes the transformation that GISMO imprints onto the image. Next, type "epar icbox" to edit the parameters of the icbox task. These are as follows: mask = output = (sz = 15.) (subraster = 100) (cursor = "") (line = "") (mode = "ql") Root name of slit mask image (e.g. S1028mask) Root name of output files (e.g. S1028) Size of alignment boxes (pixels) Size of subrasters (pixels) For the "mask" parameter, enter the root name of the output from degismo. The "output" parameter should be set to the root name (usually the name of the mask or the field) that you want the .align and .sub files to have. Next, make sure that the "sz" parameter is set to the size (in pixels) of the alignment holes as measured on the multi-slit mask image. The "subraster" parameter can be left at its default value of 100. While still in "epar" mode, type ":go" to execute icbox. One by one, the individual CCD chips of the re-mapped multi-slit mask image will be displayed, and you will be asked to mark the approximate positions of the alignment boxes. To do this, center the cursor on the box and hit "space bar". If you wish to change the minimum and maximum data values mapped into ds9, hit the "d" key. If you wish to abort, hit the "I" key. Once all the boxes on a chip have been marked (or if there are no boxes on this chip), type "q" to go to the next CCD. The task will end after cycling through all of the chips. Please make certain that each alignment box that you mark away from any edge of the GISMO slice or CCD in order to be certain that this box can be properly measured and used in the alignment process. The icbox task produces two files. If the "output" parameter of icbox was set to "S1028", these would be named "S1028.sub" and "S1028.align". The .align file will be needed during the night to carry out the alignment of the mask on the field. The .sub file is usually used for running the mask alignment process in subraster mode; this saves little time when working binned 2x2 at f/4 and is not recommended when using GISMO. Acquire images of and run icbox on the remaining GISMO masks that you intend to use that night. Set up the grating tilts for each disperser and filter combination you plan to use that night. Different masks may have different setup, depending on what you selected when you designed the masks. The simplest way to do this is to use the 'open' mask with the appropriate grating and filter combination, and the flat field screen with quartz lamps on, and then taking 'Snap' images while adjusting the tilt angle of the grating until the spectra are well centered on the detector. Suggested combinations are: Filter Grating Tilt 5700-9800 150 3.4 3600-5700 300 4.5 4800-7800 150 3 These tilts are based on past experience, but do check this in the afternoon! If you require instructions on how to turn on the flat field lamps, or on using the IMACs CamGUI, look in the other cookbooks and the user manual. Establish the appropriate flat field and wavelength arc exposure times and lamp selections for EVERY spectroscopic setup (f/4, and f/2 if you plan to use it), by adjusting exposure times and lamp selection until you are satisfied with the S/N and (lack of) saturation in these cals. You will take calibrations during the night interleaved with spectroscopic science observations, but may also take a set of cals at this point (for each mask) once you've established appropriate parameters. Finally, turn off the quartz lamp, click the mouse on the "OUT" button to take the flat field screen out of the telescope beam, and take a sequence of bias images. In the IMACS CamGUI, set Loops to the number of exposures you wish to take, ExpType to "Bias", and the Binning X and Binning Y values to whichever values you plan to use at night, and then click on the "Start" button to start the loop sequence. Although IMACS is reasonably light-tight, bias images should be taken with the dome as dark as possible. Do this for all cameras you plan to use! An appropriate catalog entry is created for you during the mask making process and is written to the SMF file. Use these entries to make an observing catalog containing all the targets you plan to observe. This includes positions for the guide stars (both principal and SH). On Sky at Night: Note: At the start of the night, make sure GISMO is inserted into the beam and clamped. Remember that GISMO may only be moved in and out horizontally. The alignment process for GISMO is very similar to that for the normal IMACS multi-slit masks. The one extra step is an IRAF task called degismo, which undoes the field rearrangement of GISMO (i.e. it produces an image that looks as though it were taken through IMACS without GISMO in place.) The normal IMACS alignment process (ialign followed by ifalign) is run on the 'de-mapped' versions of mask and field images. Have the Telescope Operator (TO) slew to the object that you wish to observe, using the catalog entry as described above. Given the appropriate catalog file position to slew to (including a guide star and SH star), the TO will first center your field using the two outer field probes and the star positions. As soon as the telescope has slewed to the position of the object, have the TO set up on a guide star and an off-axis S-H star, and begin guiding and correcting the mirror. Next take an image of the field. To do this, go to the IMACS MechGUI and make sure that the Hatch is in the "open" position, and the calibration lamps are all turned off. The Disperser must be set "f/4-Imaging"; and the GISMO mask should be 'Open' (or whatever you called the open imaging mask on your GISMO plate). Next, set the Filter to "Spectroscopic" (for maximum throughput) or use the blocking filter that you intend to use for your observations and that you already used for afternoon mask images and icbox. Now, in the IMACS CamGUI, set ExpTime to an appropriate value so that the reference stars will be visible (probably about 10-20 seconds will be sufficient), Loops to "1", ExpType to "Object", and make sure that Binning X and Binning Y is 2x2. Finally, click on the "Start" button to begin an exposure. Note that the orientation of N and E will be indicated in the QL-Tool when the exposure has read out; for GISMO data this is not very meaningful because of the re-mapping that GISMO does! Use the IRAF task degismo as before to make a 'de-mapped' version of the field image you just took. Name the output image NAMEfield, e.g., if your target is called S1028, call the output S1028field. Edit the parameters for the IRAF imacs task ialign, the parameters of which are: mask = field = align_ro = "" inter = no axis = xy sz = drange = yes z1= 1 z2= 10000 query0 = no query1 = no query2 = no query3 = yes (cursor = "") (falign = "") (mode = "ql") Image root name of slitlets (e.g. S1028mask) Image root name of field (e.g. S1028field) Alignment file ROOTNAME (e.g. S1028) Run 'geomap' interactively? Minimize which residuals? ('xy' or 'x' or 'y') Size of alignment boxes in (pixels) Set the Display range to specific values Minimum grey level to be displayed Maximum grey level to be displayed Delete stars from alignment solution? Send RA/DEC offset to TCS? Send rotation offset to TCS? Do coordinated offset? Set the "mask" parameter to the root name of the 'de-mapped' mask image created from the image taken in the afternoon; the "field" parameter should be set to the root name of image of the 'de-mapped' field image just created. Set the "align_file" parameter to the name of the .align file that you produced in the afternoon. The "inter" parameter is normally set to "no", but if you wish to interact with the alignment solution (which is performed with the iraf task "geomap"), set this parameter to "yes". Now run ialign. One by one, the expected position of the reference stars will be displayed in the "ds9" tool, and you will be asked to mark the star. Use the "space bar" to calculate the centroid, or use the "m" key to mark the position manually. If you wish to change the minimum and maximum data values mapped into ds9, hit the "d" key. If you wish to abort, hit the "I" key. Once you've marked each of the reference stars, a solution is calculated and you will be offered the chance to eliminate discrepant stars. A final solution for both a rotation offset and an offset in RA and DEC is then calculated, and you will be asked if you wish to apply these offsets. Usually it is o.k. to reply "yes". Make sure you also reply "yes" to the question of whether you want to do a coordinated offset (which moves both the telescope and the guide probes), but let the TO know before actually executing it. Now take another image of the object -- but this time with the appropriate GISMO mask selected. As before, use degismo to 'de-map' the image. Run the IRAF IMACS task ifalign on the new 'de-mapped' image. (Note the "f" in ifalign which distinguishes this task from the ialign task discussed in the previous bullet!). The parameters for ifalign are identical to those of ialign except that only one image, "mask", is requested. The ifalign task will step through each of the alignment stars showing you x and y cuts - see the standard multi-slit observing cookbook for more details. The dashed line in these plots shows the level determined for the sky. The rectangle shows the position measured for the alignment box, and the solid line displays the calculated centroid of the star. You can change any of these values (type "?" to see how to change them), but usually this is totally unnecessary. Once you are happy with the xy fits, type "q" to move on to the next alignment star. After marking all the stars, you will be offered the opportunity again to eliminate discrepant stars. Finally, new offsets are calculated and you are asked whether or not you wish to execute them. These offsets should be small, but it is usually worth executing them. You should now be ready to observe your object. In the IMACS MechGUI, set the Disperser to the grating or grism that you will use, and set the Filter to "Spectroscopic" or your desired blocking filter. In the appropriate IMACS CamGUI, set ExpTime, Loops, Binning X, and Binning Y to the desired values, and make sure you are in "Full" readout mode. To begin the exposure, click on the "Start" button. Warning: Some inconsistent behaviour has been reported with ifalign; this was likely caused by the previously reported (and now fixed) issue with the alignment script interaction with degismo. The current expectation is that BOTH ialign and ifalign should work properly, but it is worth paying attention to this - perhaps by taking a second image after running ifalign and ensuring that the objects are indeed well centered! Setting up GISMO: 1. Put the plate in GISMO and make sure it fits well. There are three small tabs cut out on the edge of the mask, which correspond to tabs on the GISMO mask holder; there is a unique rotation and flip to match the tabs up. Align the tabs carefully. You may need to trim about 1mm along one edge of the mask to make it fit; this is a known problem, due to a flaw in the mask blanks (the next batch of GISMO masks will not have this problem). The best solution for trimming the masks found so far is to use the paper cutter in the library in the support building; be SURE to trim the correct edge or you will ruin the mask! Write down the labels of each mask in the GISMO plate once you have installed it. 2. Install GISMO in IMACS and connect the power cable. 3. Power on the GISMO control electronics (in the large cabinet in the IMACS on the platform.) 4. Restart GISMO in the IMACS instrument control. 5. Add your mask labels to the GISMO gui by running in an xterm imacs setuptool. 6. Hit maskload and initialize GISMO. 7. Move GISMO into the beam. Do this only in the horizontal position. To put IMACS in the horizontal position you need to compute the rotator angle as: Rot = 134 – ZD, where the ZD = 90 – Elevation, or Rot = 44 + Elevation. (you may also use angles 180 degrees from this) There has been a problem with the feedback from the clamp which locks GISMO into place once it is inserted. Either the clamp doesn't turn green or the mask state doesn't turn green, although the insertion and clamping works fine. If this problem occurs it can probably be safely ignored, though please do document this so we can address it.